Brain tumor segmentation |
| Dana Cobzas, Karteek Popuri, Neil Birkbeck, Mark Schmidt, Martin Jagersand Albert Murtha |
ReferencesPopuri, K., Cobzas, D., Mutrtha A., and Jagersand 3D Variational Brain Tumor Segmentation using Dirichlet Priors on a Clustered Feature Set , International Journal of Computer Assisted Radiology and Surgery,2011 Popuri K., Cobzas D., Jagersand M.,Shah S.L., Murtha A. 3D variational brain tumor segmentation on a clustered feature set, SPIE Medical Imaging 2009 to appear Cobzas, D., Birkbeck, N., Schmidt, M., Jagersand, M., Murtha A. 3D Variational Brain Tumor Segmentation using a High Dimensional Feature Set, Mathematical Methods in Biomedical Image Analysis (MMBIA 2007), in conjunction with ICCV |
Demos |
Brain tumor segmentation |
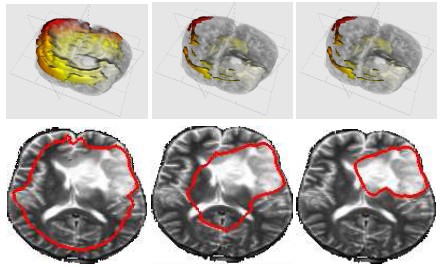
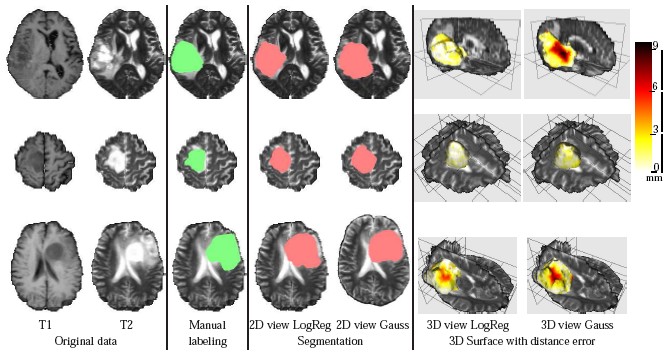
DescriptionTumor segmentation from MRI data is an important but time consuming manual task performed by medical experts. Automating this process is challenging due to high diversity in appearance of tumor tissue among different patients and, in many cases, similarity with normal tissue. One other challenge is how to make use of prior information about the appearance of normal brain. In this paper we show how to incorporate prior information into a multi-dimensional volumetric features set. Using manually segmented data we learn a statistical model for tumor and normal tissue using the same feature set.
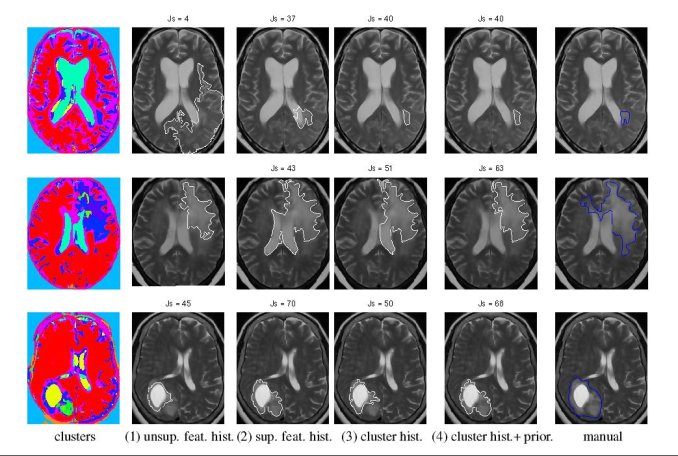
We experimented with three types of statistics - a generative Gaussian model, a discriminative Logistic Regression Model and a Parzen histogram calculated on each feature. Finally we used histogram of clustered featured and impose a Dirichlet prior to disambiguate the tumor from ventricle. The prior penalizes the clusters predominant in the ventricles from having a high probability in the tumor.
|